Today I gave a talk at DevConf.cz, which I previously gave as a keynote this past November at SeaGL, about UX and Free Software using the ChRIS project as an example. This is a blog-formatted version of that talk, although you can view a video of it here from SeaGL if you’d rather a video.
Let’s talk about a topic that is increasingly critical as time goes on, that is really important for those of us who work on free software and care really deeply about making a positive impact in the world. Let’s talk about user experience (UX) and free software, using the ChRIS Project as a case study.
What is the ChRIS Project?

The ChRIS project is an open source, free software platform developed at Boston Children’s Hospital in partnership with other organizations – Red Hat (my employer,) Boston University, the Massachusetts Open Cloud, and others.
The overarching goal of the ChRIS project is to make all of the amazing free software in the medical space more accessible and usable to researchers and practitioners in the field.

This is just a quick peek under the hood – because I’m saying ChRIS is a “platform,” but it can be unclear what exactly that means. ChRIS’ core is the backend, we call it “CUBE” – various UI are attached to that, which we’ll cover in a bit. The backend currently connects to OpenStack and OpenShift running on the Massachusetts Open Cloud (MOC.) It’s a container-based system, so the backend – which is also connected to data storage – pulls data from a medical institution, pushes the data into a series of containers that it chains together to construct a full pipeline.
Each container is a free software medical tool that performs some kind of analysis. All of them follow an input/output model – you push data into the container, it does the compute, you pull the data out of it, and pass that output on to the next container in the chain or back to up to the user via a number of front ends.
This is just a quick overview so you understand what ChRIS is. We’ll go into a little more detail later.
Who am I?

So who am I and what do I know about any of this anyway?
I’m a UX practitioner and I have been working at Red Hat for 16 years now. I specialize in working with upstream free software communities, typically embedded in those communities. ChRIS is one of the upstream projects I work in.
I’m also a long-term free software user myself. I’ve been using it since I was in high school and discovered Linux – I fell in love with it and how customizable it is and how it provides an opportunity to co-create my own computing environment.
Principles of UX and Software Freedom

From that experience and background, having worked on the UX on many free software projects over the years, I’ve come up with two simple principles of UX and software freedom.
The first one is that software freedom is critical to good UX.
The second, which I want to focus particularly here, is that good UX is critical to software freedom.

(If you are curious about the first principle, there is a talk I gave at DevConf.us some time ago that you can watch.)
3 Questions to Ask Yourself
So when we start thinking about how good UX is critical to software freedom, I want you to ask yourself these three questions:



- If a tree falls in the forest and no one is around to hear it… does it make a sound? (You’ve probably heard some form of this before.)
- If your software has features and no one can use those features… does it really have those features?
- If your software is free software, but only a few people can use it… is it really providing software freedom?
Lots of potential…
Here, in 2021, we have a wealth of free & open source technology available to us. Innovation does not require starting from scratch! For example:
- In seconds you can get containerized apps running on any system. (https://podman.io/getting-started/)
- In minutes you can deploy a Kubernetes cluster on your laptop. (https://www.redhat.com/sysadmin/kubernetes-cluster-laptop)
- In minutes you can deploy a deep learning model on Kubernetes. (https://opensource.com/article/20/9/deep-learning-model-kubernetes)
This is amazing – in free software, we’ve made so much progress in the past decade or so. You can work in any domain and startup a new software project, and all of the underlying infrastructure and plumbing you need is already available, off-the-shelf, free software licensed, for you to use.
You can focus on the bits, the new innovation, that you really care about, and avoid reinventing the wheel on the foundation stuff you need.
… and too much complication to easily realize that potential.

The problem – well, take a look at this. This is the cloud native landscape put out by the Cloud Native Computing Foundation.
I don’t mean to pick on cloud technology at all – you’ll see this level of complication in any technical domain, I think. It’s…. a little complicated, right? There’s just so much. So many platforms, tools, standards, ways of doing things.
Technologists themselves have a hard time keeping up with this.

How do we expect medical experts and clinicians to keep up with that, when even software developers have a difficult time keeping up?

The thing is, there’s a lot of potential here – there really are so many free software tools in the medical space. Some of them have been around for years.
By default, they tend to be developed and released as free software, because many are created by researchers and academic labs that want to collaborate and share.

But you know, as a medical practitioner – how do you actually make use of them? There’s a few reasons they end up being complicated to use:
- They’re often built by researchers who don’t typically have a software development background.
- They’re usually built for use in a specific study or under a specific lab environment without wider deployment in mind.
- There tends to be a lack of standardization in the tools and lack of integration between them.
- Depending on the computation involved, they may require a more sophisticated operating environment than most clinical practitioners have access to.
- There’s a high barrier to entry.
Even though these tools are free software and publically available, these aren’t tools your typical medical practitioner could pick up and start using in their practice.
Free software and hacking the Gibson

We have to remember that these are very smart people in the medical field. Neuroscientists and brain surgeons, for example. They’re smart, but they can’t “hack the Gibson.”
A good UX does not require your users to be hackers.
Unfortunately, traditionally and historically, free software has kind of required users to be hackers in order to make the best use of it.
Bridging the gap between free software and frontline usage

So how do we bridge this gap between all of this amazing free software and clinical practice, so this free software can make a positive difference in the world, so it could feasibly positively impact medical outcomes?
Good UX bridges the gap. This is why good UX is so critical to software freedom.

If your software is free software, but only a few people can use it, are you really providing software freedom?
I’m telling you no, not really. You need a good UX to be able to do that – to allow more than a few people to be able to use it, and to be able to provide software freedom.
What are these tools, anyway?
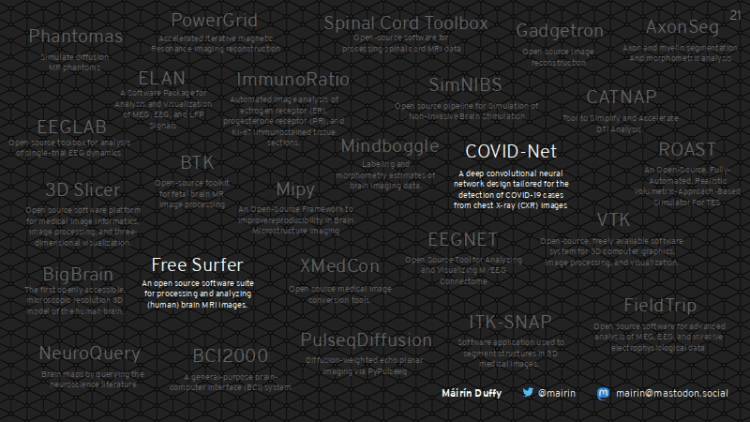
What are all of these amazing free software tools in the medical space?
This is a very quick map I made, based on a 5-10 minute survey of research papers and conference proceedings in various technology-related medical groups. These are all free software tools.
This barely scratches the surface of what is available.
I want to talk about two of them in particular today: COVID-Net and Free Surfer. They are both tools now available for use on the ChRIS platform.
Free Surfer

Freesurfer is an open source suite of tools that focuses on processing brain MRI images.
It’s been around for a long time but it’s not really in clinical use.
This is a free software tool that has a ton of potential to impact medicine. This is a screenshot of a 3D animation created in Free Surfer running on the ChRIS platform. The workflow here involved taking 2D images from an MRI, that are taken in slices across the brain. Running on ChRIS, Freesurfer constructed a 3D volume out of those flat 2D images, and then segmented the brain into all its different structures, color-coding them so you can tell them apart from one another.
How might this be used clinically? You could have a clinician who’s not sure what’s wrong with a patient. Instead of just reviewing the 2D slices, she may pan around this color coded 3D structure and notice one of the structures in the brain is larger than is typical. That might be a clue that gets the patient to a quicker diagnosis and treatment.
This is just a hypothetical example so you can see some of the potential of this free software tool.

Another example of a great free software tool in this space is COVID-Net.
This is a free software project that is developed by a company called DarwinAI in partnership with the University of Waterloo. It uses a neural network to analyze CT and Xray chest scans and provide a probability if a patient has healthy lungs, covid, or pneumomia.
It’s open source and available to the general public.
The potential here is to provide an alternative way of triaging patients when perhaps COVID test results are backed up or too slow especially during a surge in COVID cases.
These are just two projects we’ve worked with in the ChRIS project.
How do we get these tools to the frontline, though?

How do we get amazing tools like these in the frontlines, in medical institutions? How do we provide the necessary UX to bridge the gap?
Dr. Ellen Grant, who is Director of the Fetal-Neonatal Neuroimaging and Developmental Science Center at Boston Children’s Hospital, came up with a list of three basic user experience requirements these tools need for clinicians to be able to use them:
- They have to be reproducible.
- They have to be rapid.
- They have to be easy.
Requirement #1: Reproducible
First, let’s talk about reproducibility. In the medical space, you’re interacting with scientists and medical researchers trying to find evidence that supports the effectiveness of new technology or methods. So if a new method comes out – let’s say it’s a new machine learning model – and you’re reading a study showing support for its effectiveness.
If you want the best possible shot at the technique achieving a similar level of effectiveness with your own data, you’ve got to use the same version of the code, in as similar a running environment as possible as in the study. You want to eliminate any variables – like the operating environment – that might skew the output.

Here’s a screenshot of setting up Freesurfer. This is just not something we can expect medical practitioners to go through in order to reproduce an environment.

How do we make free software tools more reproducible for clinicians?
I’ll use the COVID-Net tool as an example. We worked with the COVID-Net team and they packaged it into a ChRIS plugin container. The ChRIS plugin container contains the actual code and includes a small wrapper on top with metadata and whatnot. (Here is the template for that, we call it the ChRIS cookiecutter.)
Once a tool has been containerized as a ChRIS plugin, it can run on the ChRIS platform, which gives you a number of UX benefits including reproducibility. A clinician can just pick that tool from a list of tools from within the ChRIS UI, push their data to it, and get the results, and ChRIS manages the rest.

Taking a few steps back – we have a broader vision for reproducibility via ChRIS here.
This is a screenshot of a prototype of what we call the ChRIS Store.
We envision making all of these amazing free software medical tools as easy to install and run on top of ChRIS as it is to install and run apps on a phone from an app store. So this is an example of a tool containerized for ChRIS – you’d be able to take a look at the tool in the ChRIS Store, deploy it to your ChRIS server, and use it in your analysis.
Even if a tool is a little hard to install, run, and reproduce in the same exact way on its own, for the small cost of packaging and pushing it into the ChRIS plugin ecosystem, it becomes much easier to share, deploy, and reproduce that tool across different ChRIS servers.

Instead of requiring medical researchers and practitioners need to use the linux terminal, to compile code, and to set up environments with exact specifications, we envision them being able to browse through these tools, like in an app store, and be able to easily run them on their ChRIS server. That would mean they would get much more reproducibility out of these tools.
Requirement #2: Rapid

The second requirement from Dr. Grant is rapidness. These tools need to be quick. Why?
Well, for example, we’re still in a pandemic right now. As COVID cases surge, hospitals run out of capacity and need to turn over beds quickly. Computations that take hours or days to run will just not be used by clinicians, who do not have that kind of time. So the tools need to be fast.
Or for a non-pandemic case… you might have a patient who needs to travel far for specialized care – if results could come back in minutes, it could save a sick patient from having to stay in a hotel away from home and wait days for results and to move forward in their treatment.

Some of these computations take a long time, so couldn’t we throw some computing power at them to get the results back quicker?
ChRIS lays the foundation that will enable you to do that. ChRIS can run or orchestrate workloads on a single system, HPC, or a cloud, or across those combined. You can get really rapid results, and ChRIS gives you all the basic infrastructure to do it, so individual organizations don’t have to figure out how to set this up on their own from scratch.

For example – this is a screenshot of the ChRIS UI – it shows how you build these pipelines or analyses in ChRIS. The full pipeline is represented by the graph on the left, and each of the circles or “nodes” on the graph is a container running on ChRIS. Each of these containers is running a free software tool that was containerized for ChRIS.
The blue highlighted container in the graph is running Freesurfer. In this particular pipeline, ChRIS has spun up different copies of the same chain of containers as to run probably on different pieces of the data output by that blue Freesurfer node.
You can get this kind of orchestration and computing power just based on the infrastructure you get from ChRIS.

This is a diagram to show another view of it.
You have the ChRIS store at the top with a plugin (P) getting loaded into the ChRIS Backend.
You have the data source – typically a hospital PACS server with medical imaging data, and image data (I).
ChRIS orchestrates the movement of the data and deployment of these containers into different computing environments – maybe one of these here is an external cloud, for example. ChRIS retrieves the data from the data source and pushes it into the containers, and retrieves the container pipeline’s output and stores it, presenting it to the end user in the UI. Again, each one of those containers represents a node on the pipeline graph we saw in the previous slide, and the same pipeline can consist of nodes running in different computing environments.

One of those compute environments that ChRIS utilizes today is the Massachusetts Open Cloud.
This is Dr. Orran Krieger, he is the principal investigator for the Massachusetts Open Cloud at Boston University.
The MOC is a publicly-owned, non-commercial cloud. They collaborate with the ChRIS project, and we have a test deployment of ChRIS that we are using for COVID-Net user testing right now that runs on top of some of the powerpc hardware in the MOC.
The MOC partnership is another way we are looking to make rapid compute in a large cloud deployment accessible for medical institutions – a publicly-owned cloud like the MOC means institutions will not have to sign over their rights to a commercial, proprietary cloud who might not have their best interests at heart.
Requirement #3: Easy
Finally, the last UX requirement we have from Dr. Grant is “easy.”
What we’ve done in the ChRIS project is to create and assemble all of the infrastructure and plumbing needed to connect to powerful computing infrastructures for rapid compute. And we’ve created a container-based structure and are working on creating an ecosystem where all of these great free software tools are easily deployable and reproducible and you can get the same exact version and env as studied by researchers showing evidence of effectiveness.
One of the many visions we have for this: a medical researcher could attend a conference, learn about a new tool, and while sitting in the audience (perhaps by scanning a QR code provided by the presenters) access the same tool being presented in the ChRIS store. They could potentially deploy to their own ChRIS on their own data to try it out, same day.
This all needs to be reproducible, and it needs to be easy. I’m going to show you some screenshots of the ChRIS and COVID-Net UIs we’ve built in making running and working with these tools easier.

This is an example of the ChRIS feed list in the Core ChRIS UI. Each of these feeds (what we call custom pipelines) is running on ChRIS. Each pipeline is essentially a composition of various containerized free software tools chained together in an end to end workflow, kicked off with a specific set of data that is pushed through and transformed along the way.
This UI is not geared at clinicians, but is more aimed at researchers with some knowledge of the types of transformations the tools create in the data – for example, brain segmentation – who want to create compositions of different tools to explore the data. They would compose these pipelines in this interface, experiment with them, and once they have created one they have tested and believe is effective, they can save it and reuse it over and over on different data sets.

While you are creating this pipeline, or if you are looking to add on to a pre-existing workflow, you can add additional “nodes” – which are containers running a particular free software tool inside – using this interface. You can see the list of available tools in the dialog there.

As you add nodes to your pipeline, they run right away. This is a view of a specific pipeline, and you can see the node container highlighted in blue here has a status display on the bottom showing that it is currently still computing. When the output is ready, it appears down there as well, per-node, and it syncs the data out and passes it on to the next node to start working on.
Again, this is an interface geared towards a researcher with familiarity analyzing radiological images – but not necessarily the skill set to compile and run them from scratch on the command line. This allows them to select the tools and bring them into a larger integrated analysis pipeline, to experiment with the types of output they get and try the same analysis out on different data sets to test it. They are more likely looking at broad data sets to see trends across them.
A practicing clinician needs vastly simplified interfaces compared to this. They aren’t inventing these pipelines – they are consuming them for a very specific patient image, to see if a specific patient has COVID, for example.

As we collaborate with the COVID-Net team, we are focused on creating a single-purpose UI that used just one specific pipeline – the COVID-Net analysis pipeline – and could allow a clinician to simply select the patient image, click and go, and get the predictive analysis results.
The first step in our collaboration was containerizing the COVID-Net tool as a ChRIS plugin. That took just a few days.
Then together over this past summer, in maybe 2-3 months, we built this very streamlined UI aimed at just this specific case of a clinician running the COVID-Net prediction on a patient lung scan and getting the results back. Underneath this UI, is a pipeline, just like the one we just looked at in the core UI – but clinicians will never see that pipeline underneath – it’ll just be working silently in the background for them.
The user simply types in a patient MRN – medical record number – to look up the scans for that patient at the top of the screen, selects the scans they want to submit, and hits analyze. Underneath that data gets pushed into a new COVID-Net pipeline.

They’ll get the analysis results back after just a minute or two, and it looks like this. These are predictive analyses – so here the COVID-Net model believes this patient has about a 75% chance of having normal, healthy lungs and around a 25% or so chance of having COVID.
If they would like to explore this a little further, maybe confirm on the scan themselves to double check the model, they can click on the view button and pull up a full radiology viewer.

Using this viewer, you can take a closer look at the scan, pan, zoom, etc. – all the basic functionality a radiology viewer has.
This is an example of the model we see for ChRIS to provide simplified, easy ways of accessing the rapid compute and reproducible tool workflows we talked about: Standing up streamlined, focused interfaces on top of the ChRIS backend – which provides the platform, plumbing, tooling to quickly stand up a new UI – so clinicians don’t have to develop their own workflows, they can consume tested and vetted workflows created by experts in the medical data analysis field.
To sum it all up –

This is how we are working to meet these three core UX requirements for frontline medical use.
We’re looking to make these free software tools reproducible using the ChRIS container model, rapid by providing access to better computing power, and easy by enabling the development of custom streamlined interfaces to access the tools in a more consumable way.

In other words, the main requirement for these free software tools to get into the hands of front line medical workers is a great user experience.

Generally, for free software to matter, for us to make a difference in the world, for users to be able to enjoy software freedom – we have to provide a great user experience so they can access it.

So in review – the two principals of software freedom and UX:
- Software freedom is critical to good UX.
- Good UX is critical to software freedom.



